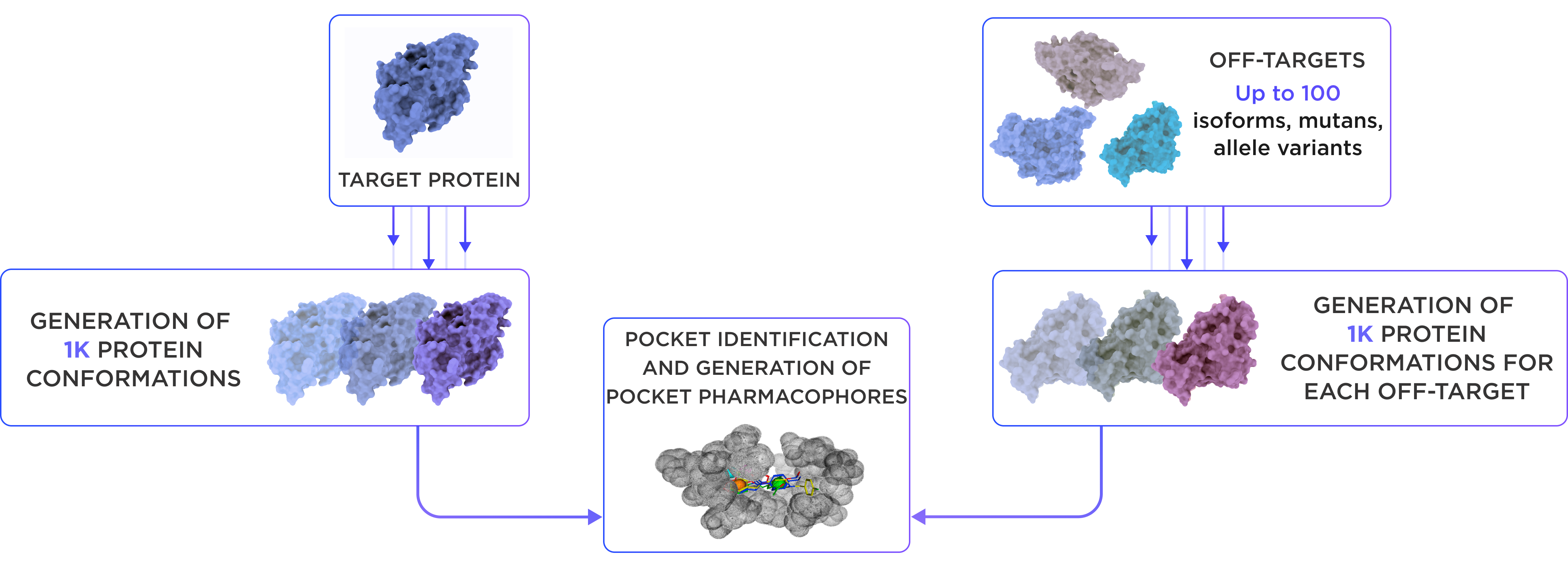
The workflow process begins with generation of 1000 conformations for target and each off-target protein is proceeded. Then all-atom MD simulations of targets and off-targets are processed using a proprietary clustering algorithm to extract representative protein conformations, accounting for overall flexibility and binding pocket dynamics. In the next step pocket identification and generation of pocket pharmacophores is conducted.
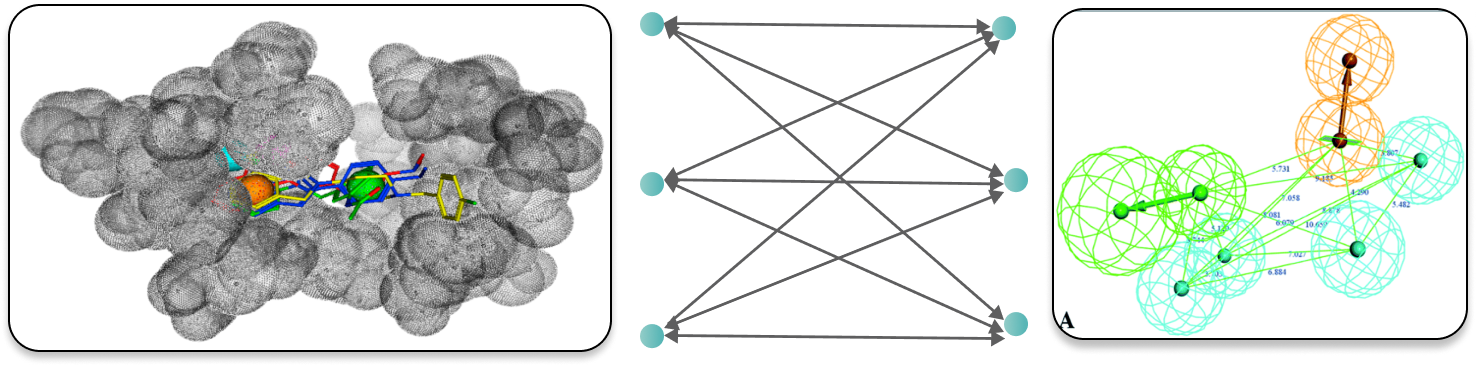
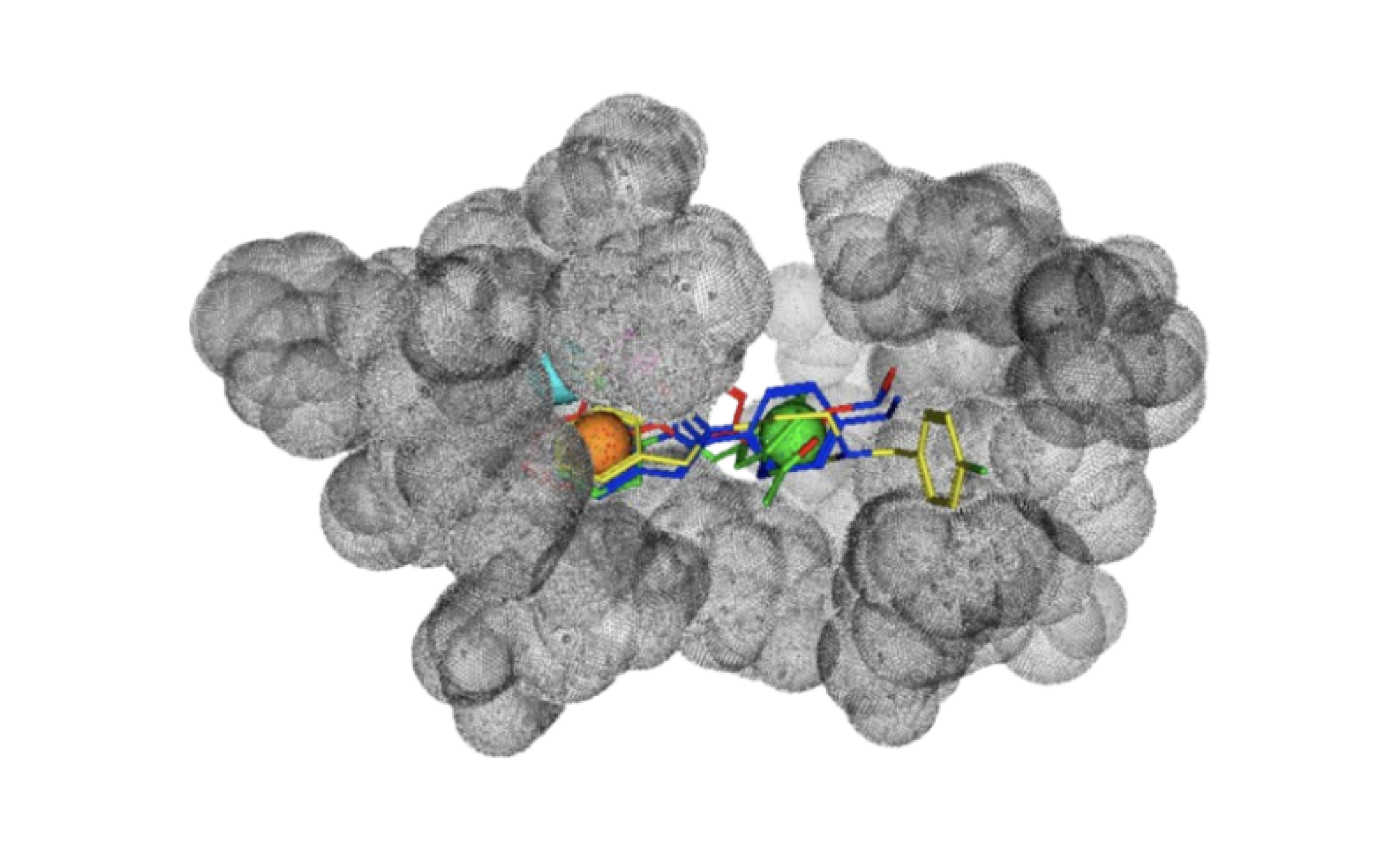
These pocket pharmacophores are superimposed, aligned and analyzed by our proprietary feature detection algorithm, which emphasises the features that are specific to the target protein. This results in a "differential pharmacophore" that represents both structural and dynamical distinctive features of the binding pocket in the target protein.
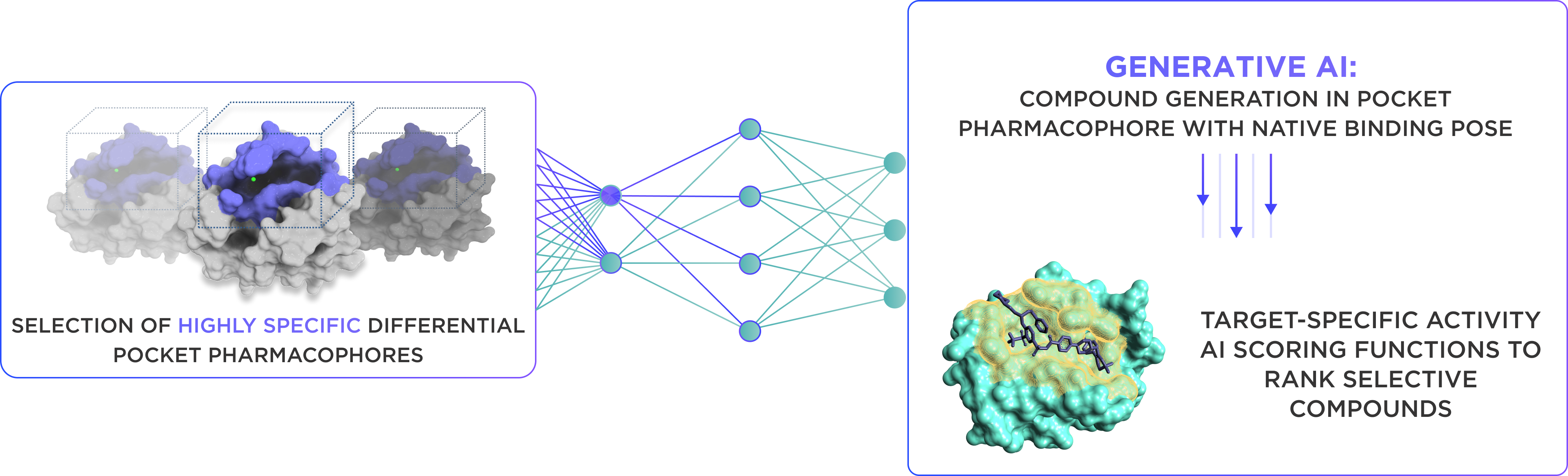
At the next stage selection of highly-specific differential pocket pharmacophores is proceeded. The generative AI model based on molecular transformer neural network predicts 3D ligand conformers that match the differential pharmacophore of the binding pocket. We used proprietary data augmentation algorithm for generating 20M protein ligand complexes for training.
This workflow allows to evaluate selectivity of compound against up to 100 off-targets simultaneously, which is crucial for drug design and the prevention of side effects.