2
novel allosteric pockets targeted
1.4M
focused library
screened
4
potent hits out of
204 candidates
18x
interferon induction by
lead-like compound
01/ Background
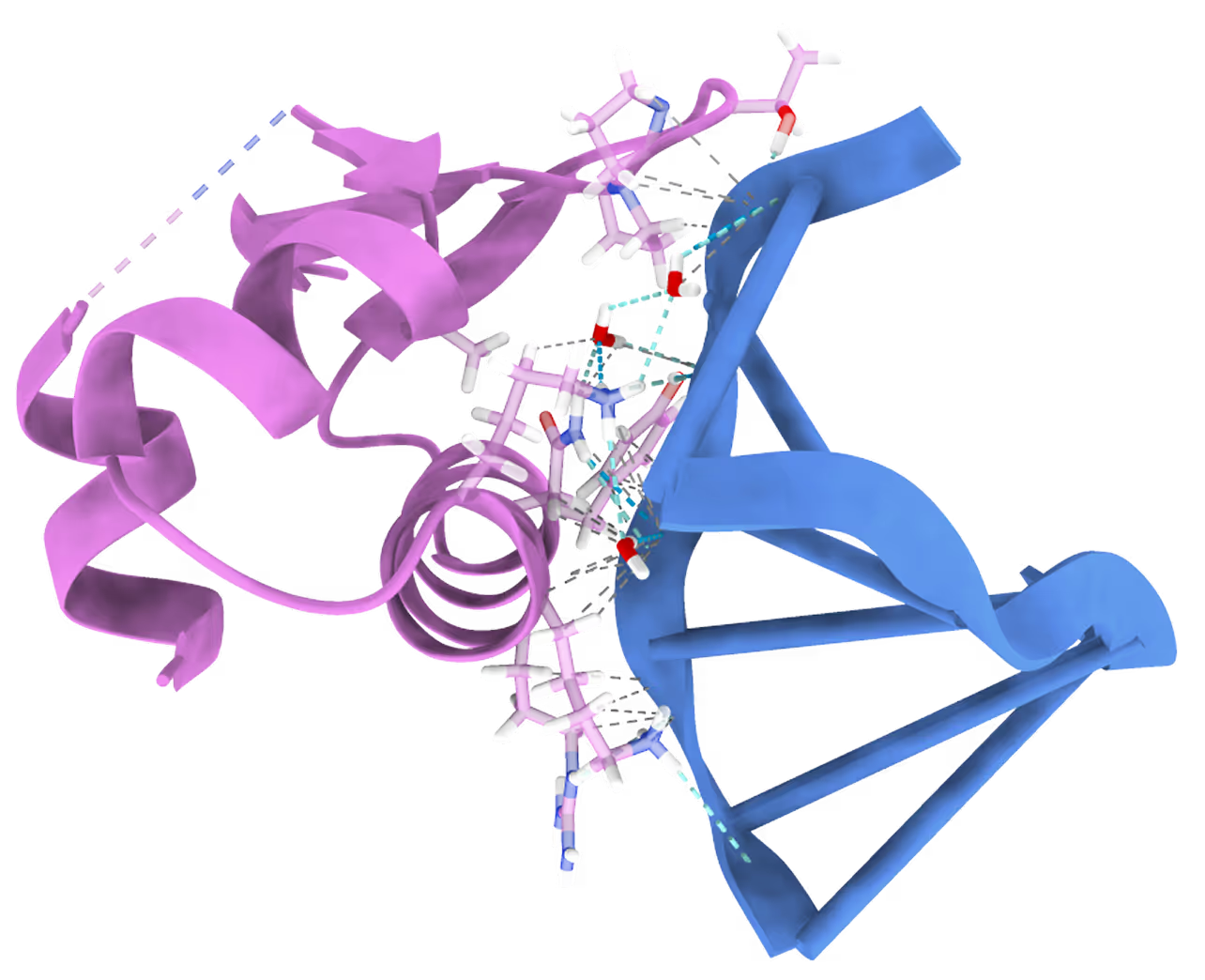
- The goal is to design an allosteric inhibitor for RNA-binding protein ADAR1 and avoid off-target effects with ADAR2.
- Only a few known allosteric inhibitors exist.
- Known allosteric pockets are poorly druggable.
02/ Methodology
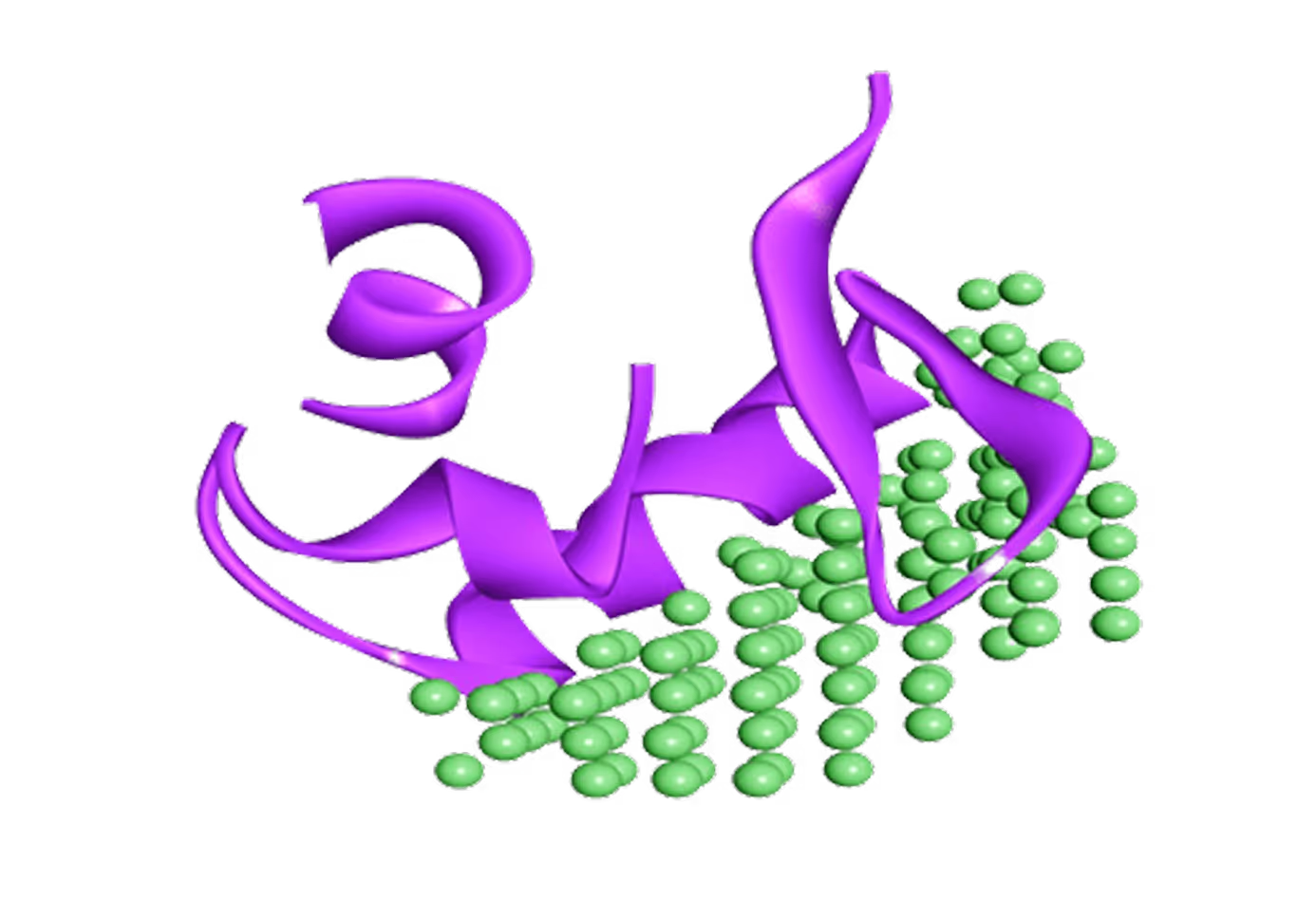
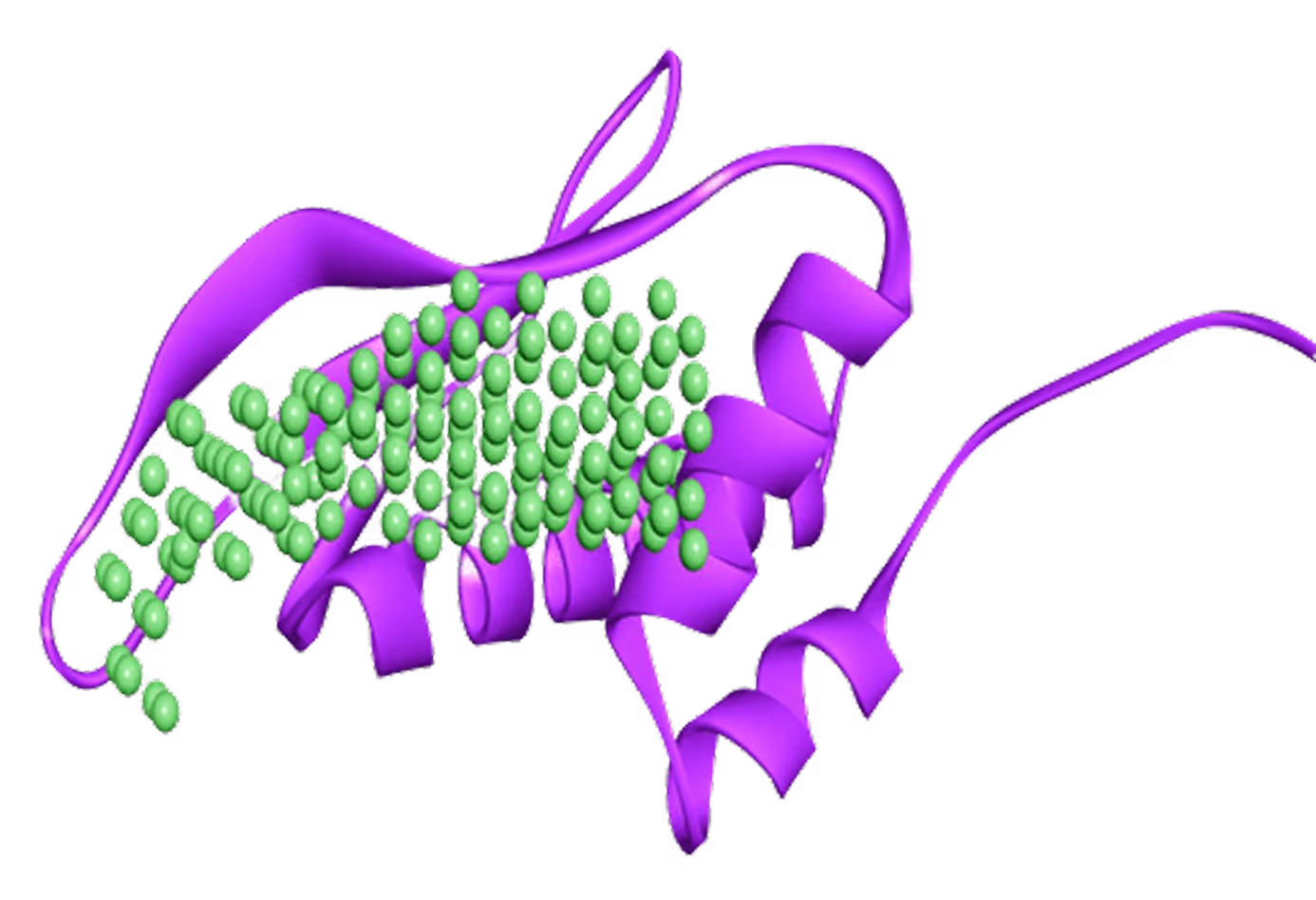
- 2 novel allosteric pockets identified by
Receptor.AI's proprietary pocket detection AI model. - Virtual screening performed for 1.4 M focused library.
- 1000 ranked compounds prioritized.
- 209 compounds subjected to in vitro validation after
AI-guided hit candidates selection. - Experimental validation performed using a
high throughput p110 knockout cell-based assay. - Hit compounds confirmed by the dose-response analysis.
03/ Results
- Criteria for a hit compound was established as a
5x fold increase in interferon induction compared to the control. - Desirable outcome to surpass the efficacy of siRNA alternatives was a 10x fold increase.
- 4 hit compounds with interferon inducing activities were identified.
- Lead-like compound with 18x fold interferon induction.
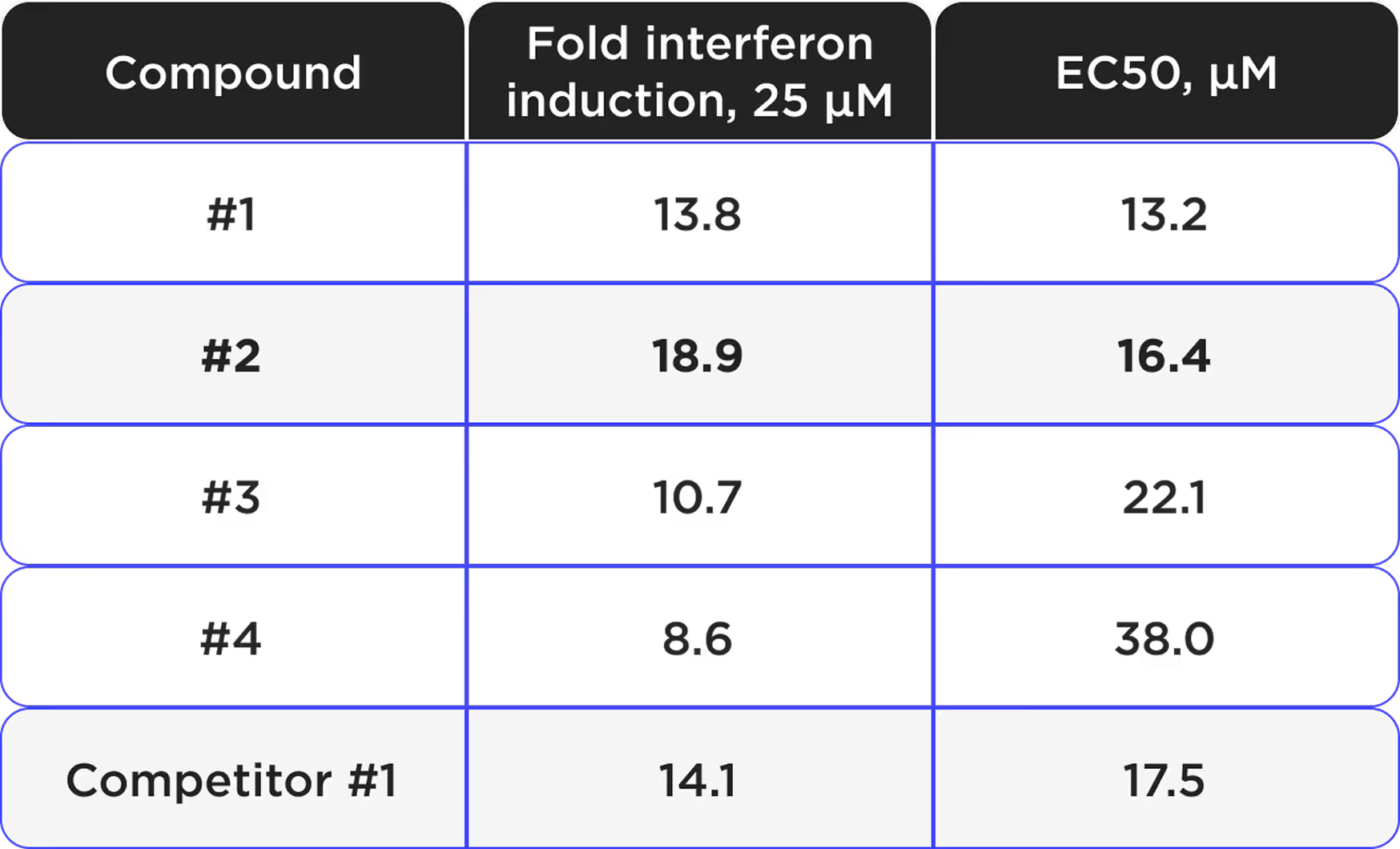
*Hit compounds identified
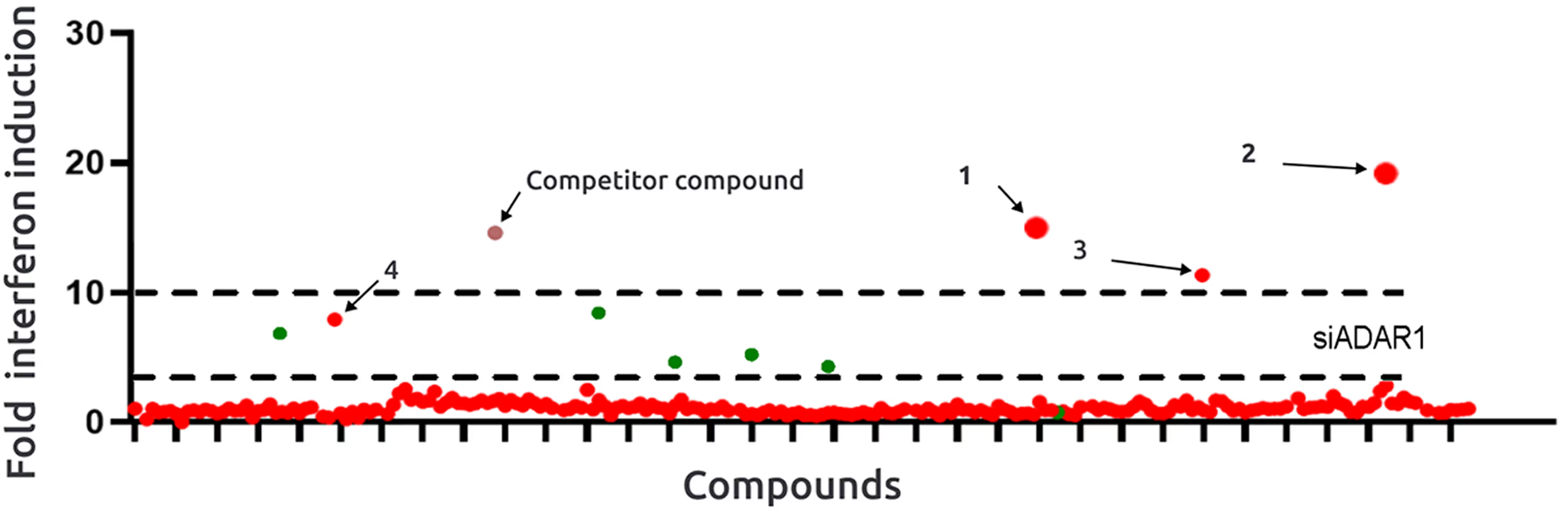
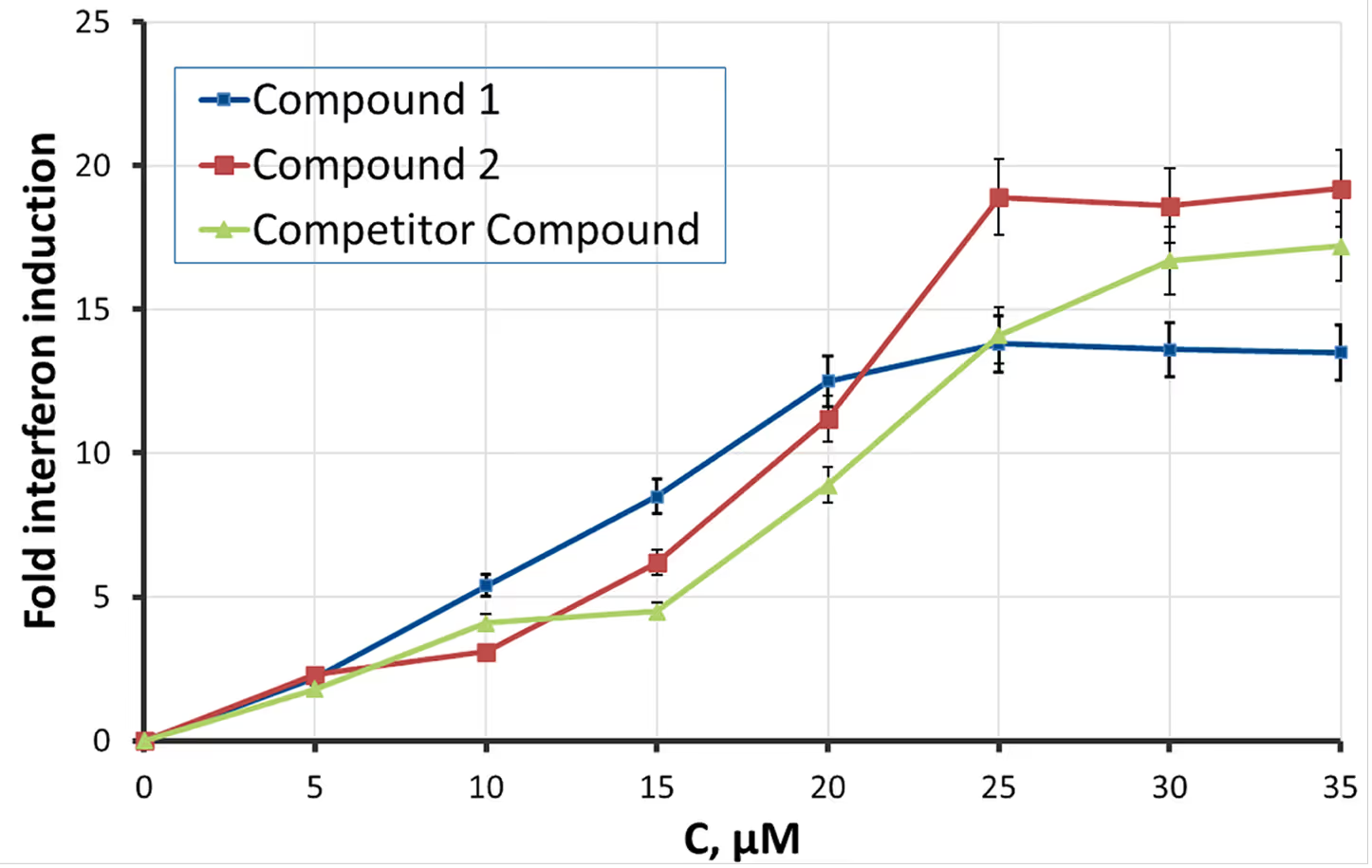
- 2 hits exhibit comparable or superior maximal interferon induction with lower EC50 in comparison to a competing compound.
- This was achieved on a 2.5x smaller screening library
(209 against 500 for competitors). - Active scaffolds have been selected for further
series expansion and optimization.