Beyond the Benchmark: Rethinking Molecular Docking Accuracy
Announcement



Full Text
What if the molecular docking accuracy we’ve trusted for decades is far lower under real-world conditions than most benchmarks suggest?
A recent study from Receptor.AI takes a critical look at how docking truly performs outside idealized test sets. Using the PLINDER-MLSB benchmark, which simulates realistic scenarios with unbound and predicted protein structures, the results are striking: even the best ML-based method, ArtiDock, achieves only ~18% success when both geometric and chemical validity are enforced, while classical tools perform significantly worse.
Even when all docking methods are combined into a single ensemble, which theoretically chooses the best pose for each target, it achieves only about 35% accuracy..
See the graph below for the complete benchmark results.
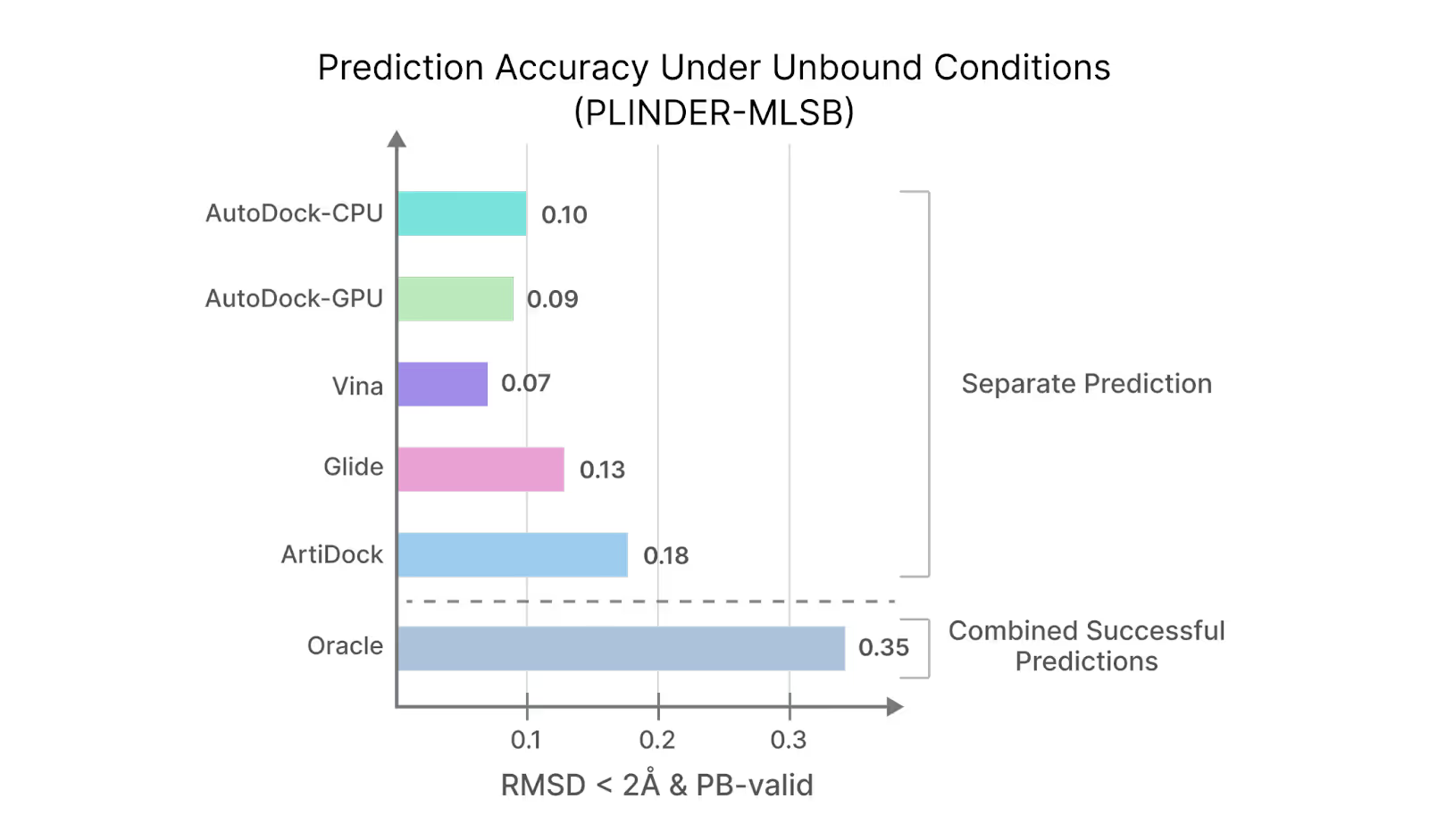
These findings challenge long-standing assumptions about docking reliability and explain why so many “good-looking” poses fail in downstream validation.
🧩 Key takeaways:
- Classical docking accuracy collapses under realistic conditions, a reminder that docking is better viewed as a statistical filter rather than a precision predictor.
- Protein rigidity, neglected solvent effects, and misassigned protonation states remain major sources of error.
- Scoring functions are still the Achilles’ heel of docking pipelines.
- Hybrid and ensemble approaches, such as Receptor.AI’s QuorumMap, show promise by combining multiple docking engines (AutoDock, Glide, Vina, ArtiDock) and applying active learning to explore chemical space more intelligently.
⚙️ ArtiDock itself stands out for its computational efficiency, being 2–3× more computationally efficient than AutoDock-GPU. Yet even this next-generation method alone cannot achieve the level of precision required for complex, real-world scenarios.
Docking doesn’t need to be replaced; it needs to evolve. When grounded in realistic biology and guided by AI, it can finally deliver on its original promise.

