Introducing the Receptor.AI ADMET Model Family for Scalable and Accurate Predictions
Introducing the Receptor.AI ADMET Model Family for Scalable and Accurate Predictions
PARTNERSHIP
Announcement



Summary
Full Text
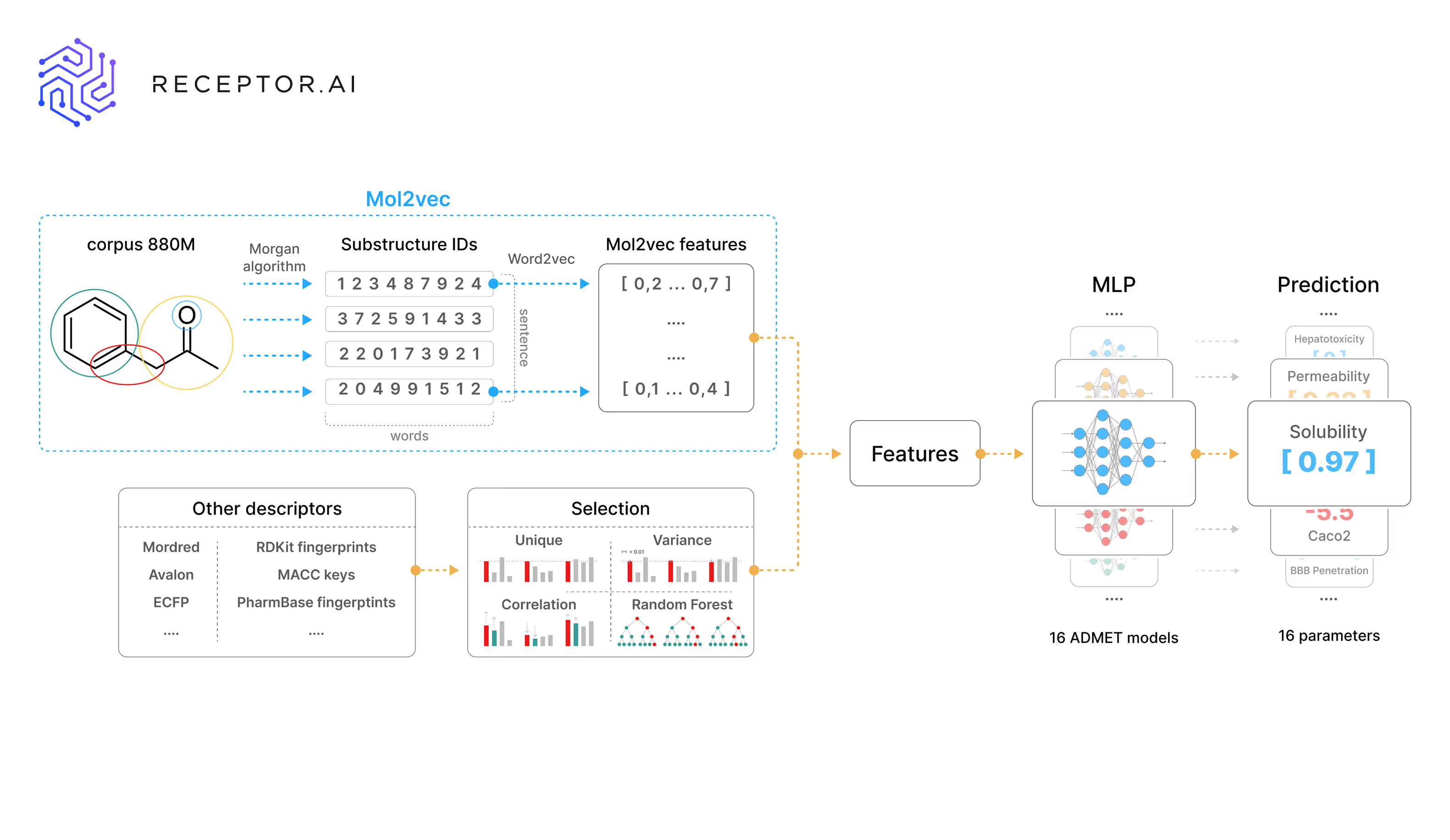
Our preprint on the new Receptor.AI ADMET prediction model family is now available. It covers the full methodology and benchmarking results behind four model variants, each designed for specific applications. These range from high-throughput Mol2Vec-only models suitable for large-scale screening to descriptor-enriched versions optimized for maximum accuracy in focused compound profiling.
The approach leverages a hybrid molecular representation, combining unsupervised Mol2Vec embeddings trained on nearly 900 million compounds from ZINC20 with curated descriptor augmentation. While many recent models rely on a single representation type, such as GNNs or transformer-based embeddings, we show that integrating chemically informed context vectors with classical descriptors consistently outperforms more complex architectures.
The models predict 40 endpoints and have been benchmarked across 16 ADMET tasks from the TDC, achieving first-place ranking on 10 endpoints. This is the best top-ranking performance reported to date, with particularly strong results on challenging endpoints including DILI, hERG, and CYP450. The models outperformed Chemprop, DeepPurpose, ZairaChem, and GNN-based methods such as MapLight.
Our model offers a unique combination of high accuracy and light-weight architecture. It exceeds in benchmarks while remaining fast, scalable, and easy to integrate into drug discovery workflows.


