

Available from Reaxense
This protein is integrated into the Receptor.AI ecosystem as a prospective target with high therapeutic potential. We performed a comprehensive characterization of Kelch-like protein 12 including:
1. LLM-powered literature research
Our custom-tailored LLM extracted and formalized all relevant information about the protein from a large set of structured and unstructured data sources and stored it in the form of a Knowledge Graph. This comprehensive analysis allowed us to gain insight into Kelch-like protein 12 therapeutic significance, existing small molecule ligands, relevant off-targets, and protein-protein interactions.
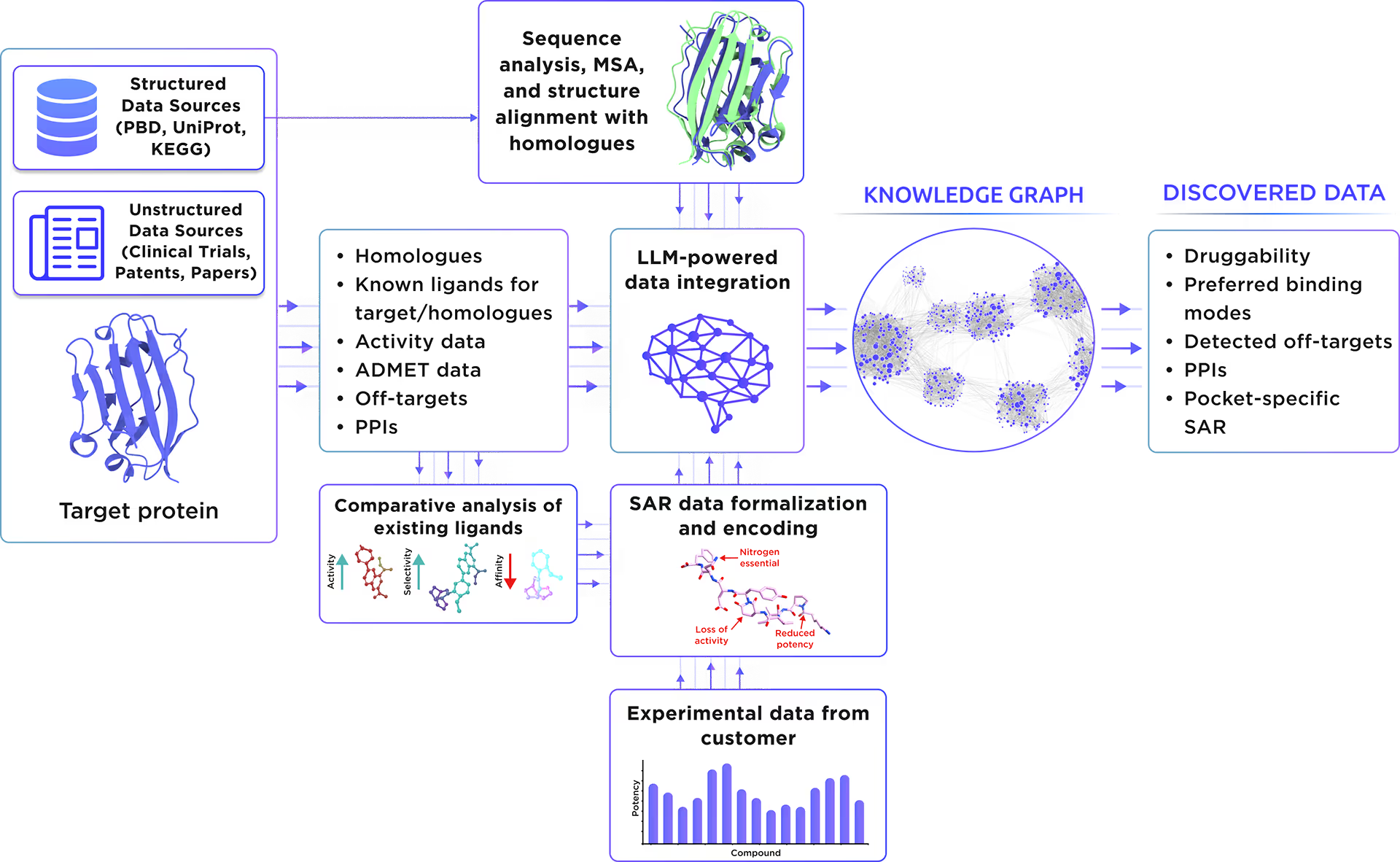
Fig. 1. Preliminary target research workflow
2. AI-Driven Conformational Ensemble Generation
Starting from the initial protein structure, we employed advanced AI algorithms to predict alternative functional states of Kelch-like protein 12, including large-scale conformational changes along "soft" collective coordinates. Through molecular simulations with AI-enhanced sampling and trajectory clustering, we explored the broad conformational space of the protein and identified its representative structures. Utilizing diffusion-based AI models and active learning AutoML, we generated a statistically robust ensemble of equilibrium protein conformations that capture the receptor's full dynamic behavior, providing a robust foundation for accurate structure-based drug design.
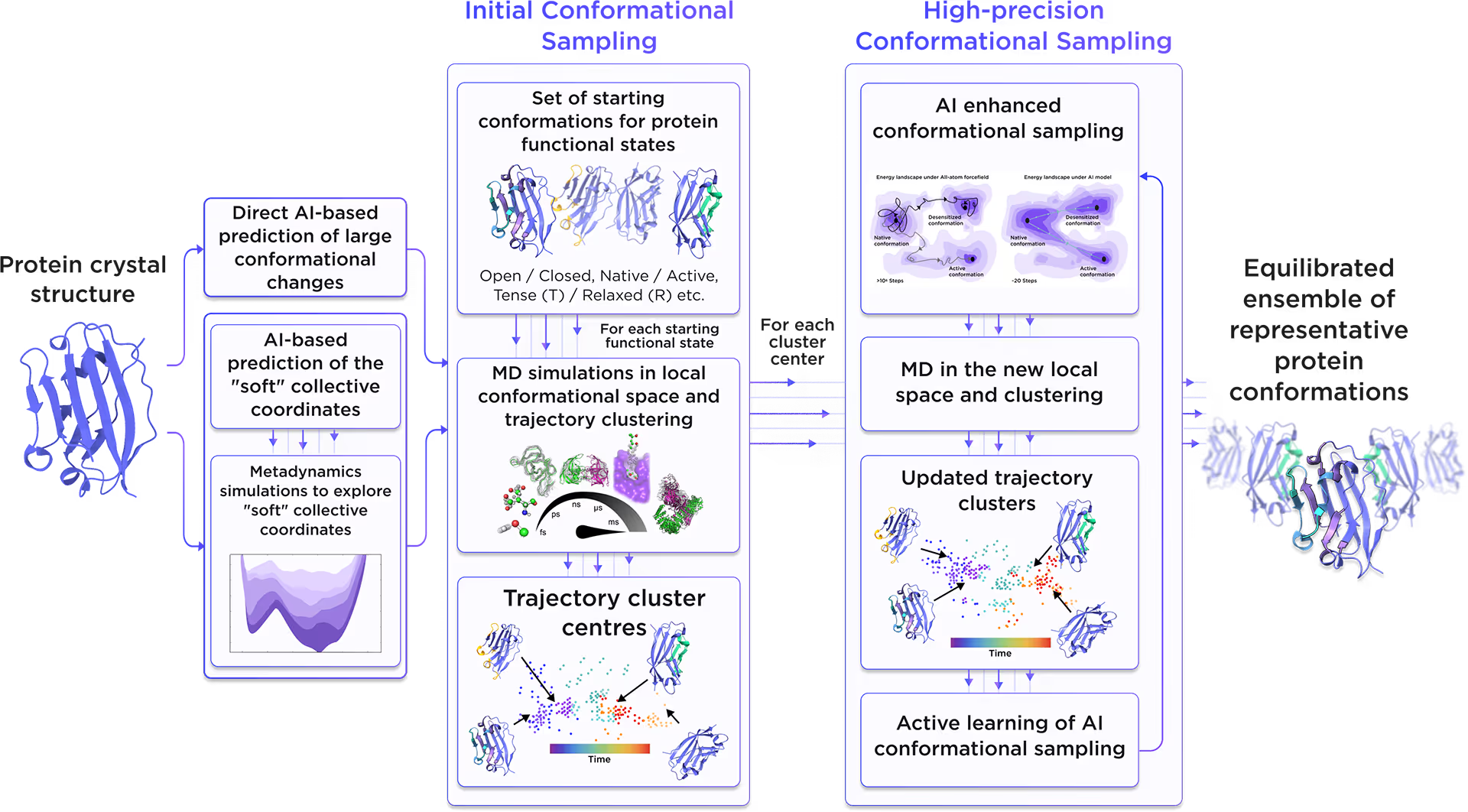
Fig. 2. AI-powered molecular dynamics simulations workflow
3. Binding pockets identification and characterization
We employed the AI-based pocket prediction module to discover orthosteric, allosteric, hidden, and cryptic binding pockets on the protein’s surface. Our technique integrates the LLM-driven literature search and structure-aware ensemble-based pocket detection algorithm that utilizes previously established protein dynamics. Tentative pockets are then subject to AI scoring and ranking with simultaneous detection of false positives. In the final step, the AI model assesses the druggability of each pocket enabling a comprehensive selection of the most promising pockets for further targeting.
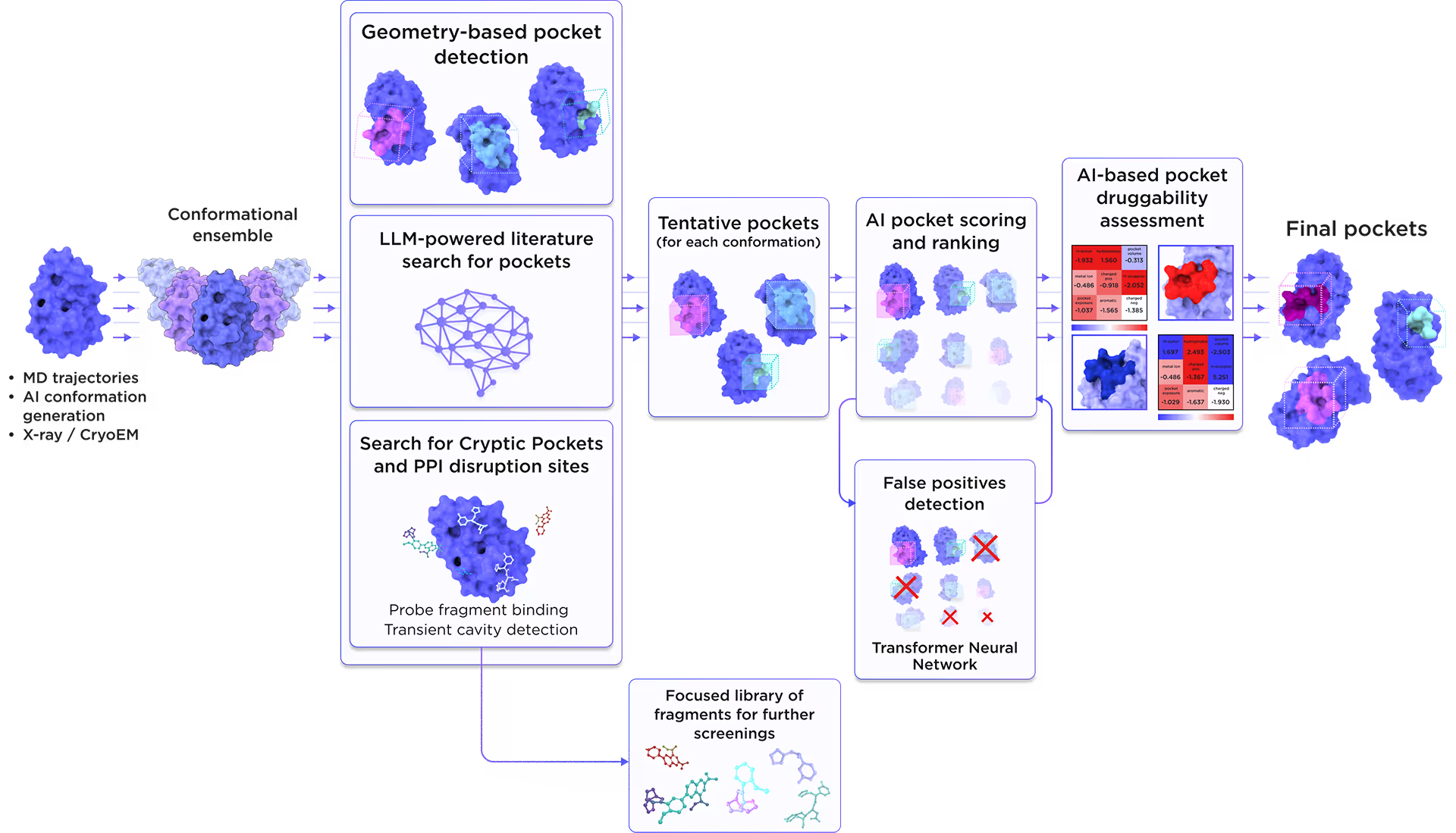
Fig. 3. AI-based binding pocket detection workflow
4. AI-Powered Virtual Screening
Our ecosystem is equipped to perform AI-driven virtual screening on Kelch-like protein 12. With access to a vast chemical space and cutting-edge AI docking algorithms, we can rapidly and reliably predict the most promising, novel, diverse, potent, and safe small molecule ligands of Kelch-like protein 12. This approach allows us to achieve an excellent hit rate and to identify compounds ready for advanced lead discovery and optimization.
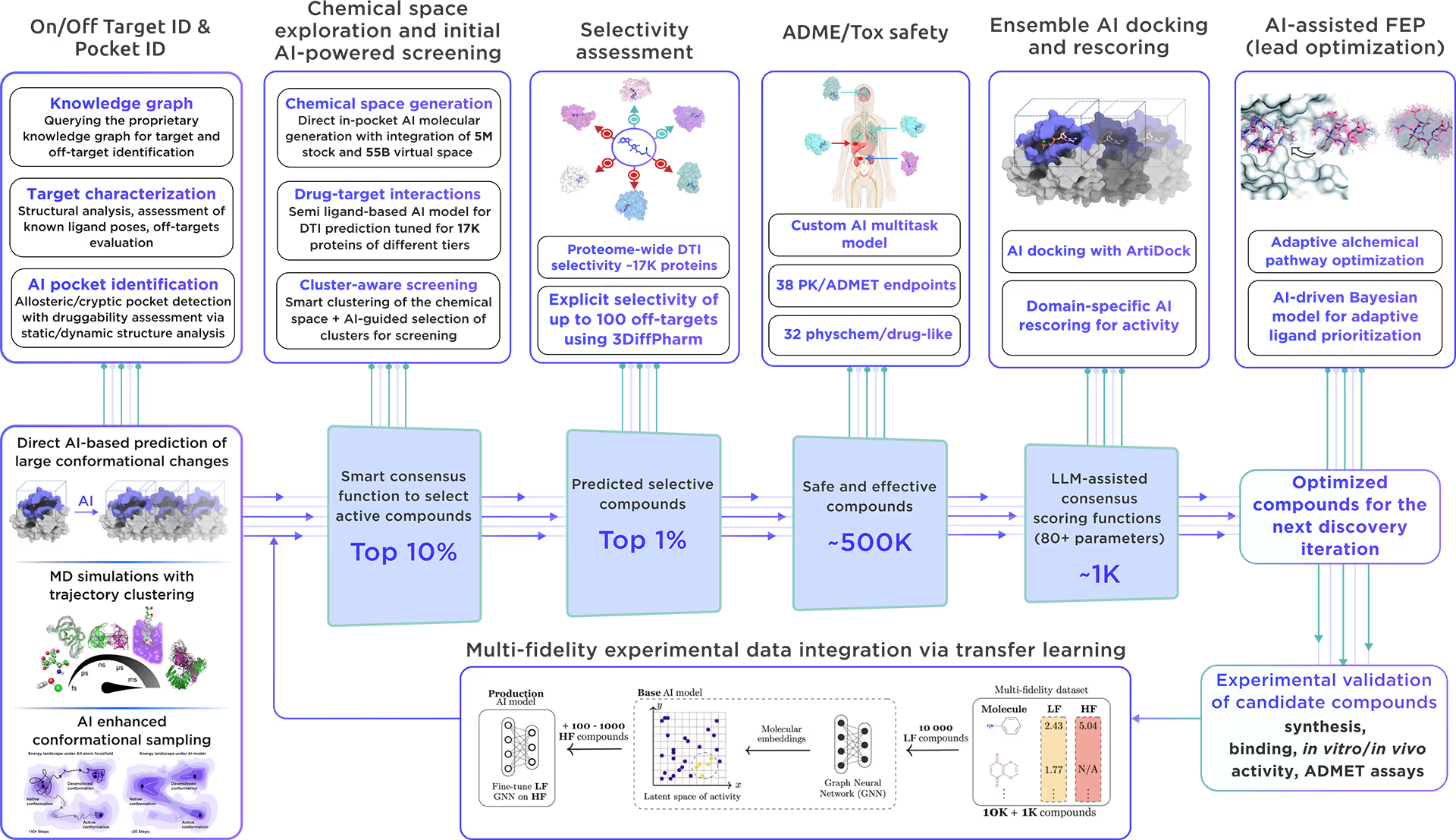
Fig. 4. The screening workflow of Receptor.AI
Receptor.AI, in partnership with Reaxense, developed a next-generation technology for on-demand focused library design to enable extensive target exploration.
The focused library for Kelch-like protein 12 includes a list of the most effective modulators, each annotated with 38 ADME-Tox and 32 physicochemical and drug-likeness parameters. Furthermore, each compound is shown with its optimal docking poses, affinity scores, and activity scores, offering a detailed summary.
Kelch-like protein 12
partner:
Reaxense
upacc:
Q53G59
UPID:
KLH12_HUMAN
Alternative names:
CUL3-interacting protein 1; DKIR homolog
Alternative UPACC:
Q53G59; A6NEN8; B7Z7B8; Q9HBX5
Background:
Kelch-like protein 12 (KLHL12), also known as CUL3-interacting protein 1 or DKIR homolog, plays a pivotal role in cellular processes. It serves as a substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin ligase complex, crucial for negative regulation of the Wnt signaling pathway and ER-Golgi transport. The BCR(KLHL12) complex is instrumental in ER-Golgi transport by regulating COPII coats size, essential for collagen export and embryonic stem cells division. It mediates monoubiquitination of SEC31, facilitating neural crest specification and collagen export. Additionally, it negatively regulates the Wnt signaling pathway through ubiquitination and proteolysis of DVL3.
Therapeutic significance:
Understanding the role of Kelch-like protein 12 could open doors to potential therapeutic strategies.