2
novel subpockets
targeted
200K
focused library
screened
18
structurally diverse hit compounds
1.6 μM
IC50 of the best compound
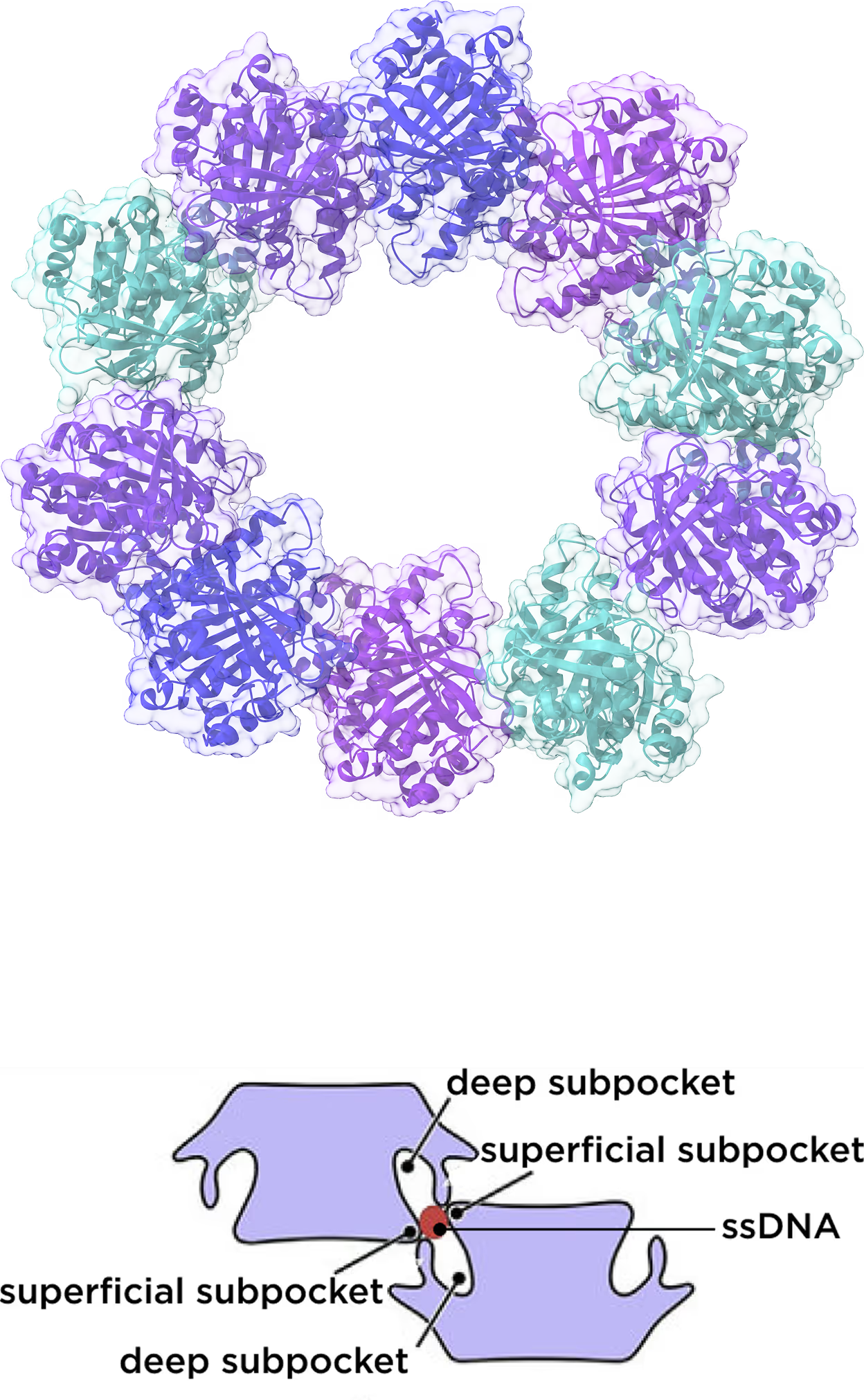
with DNA binding scheme
01/ Background
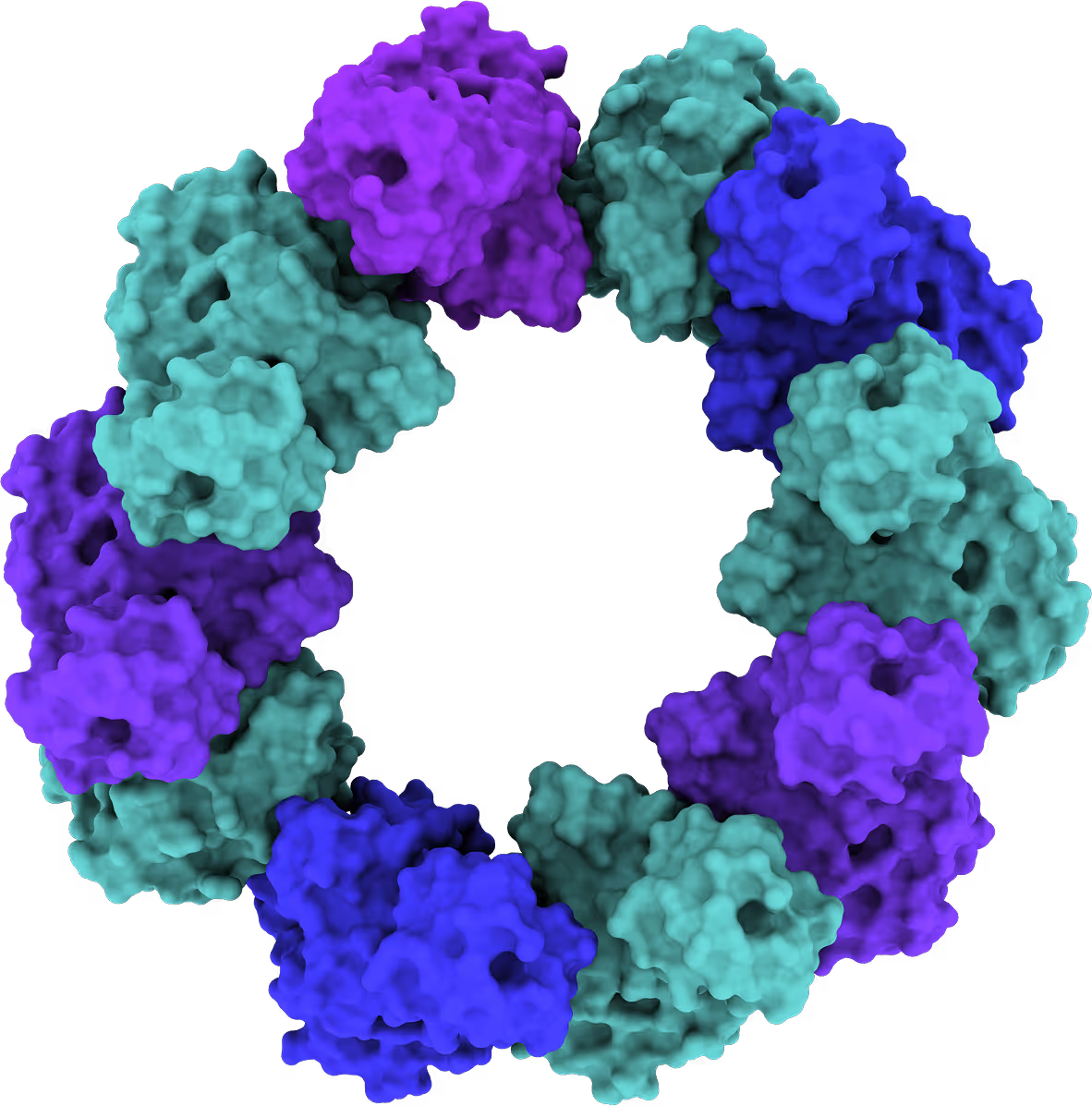
- The target is a DNA reparation and recombination protein.
- The goal is to design a competitive inhibitor with high affinity against ssDNA binding site (tough competition for binding).
- The pocket is hard-to-drug: large and highly charged.
02/ Methodology
- AI-powered MD and conformational ensemble generation.
- Hybrid pocket ID (geometric- + AI-based).
- Focused library generation (pharmacophore-/substructure-based).
- AI-driven virtual screening.
- Fluorescence polarization competition assay to identify hits.
- Dose responding assay to confirm initial hit compounds.
03/ Workflow
- Identified and targeted 2 subpockets in one iteration.
- Deep subpocket targeted with 100K focused library based on DNA-like pharmacophore generated from 8M stock.
- Superficial subpocket targeted with 100K focused library of compounds with acceptor groups generated from 8M stock.
04/ Results
- 287 hit candidates tested in vitro.
- 18 structurally diverse hit compounds with IC50 < 10 μM identified.
- 10 hits confirmed by dose responding assay.
- The top hit shows IC50 ~1.6 μM.
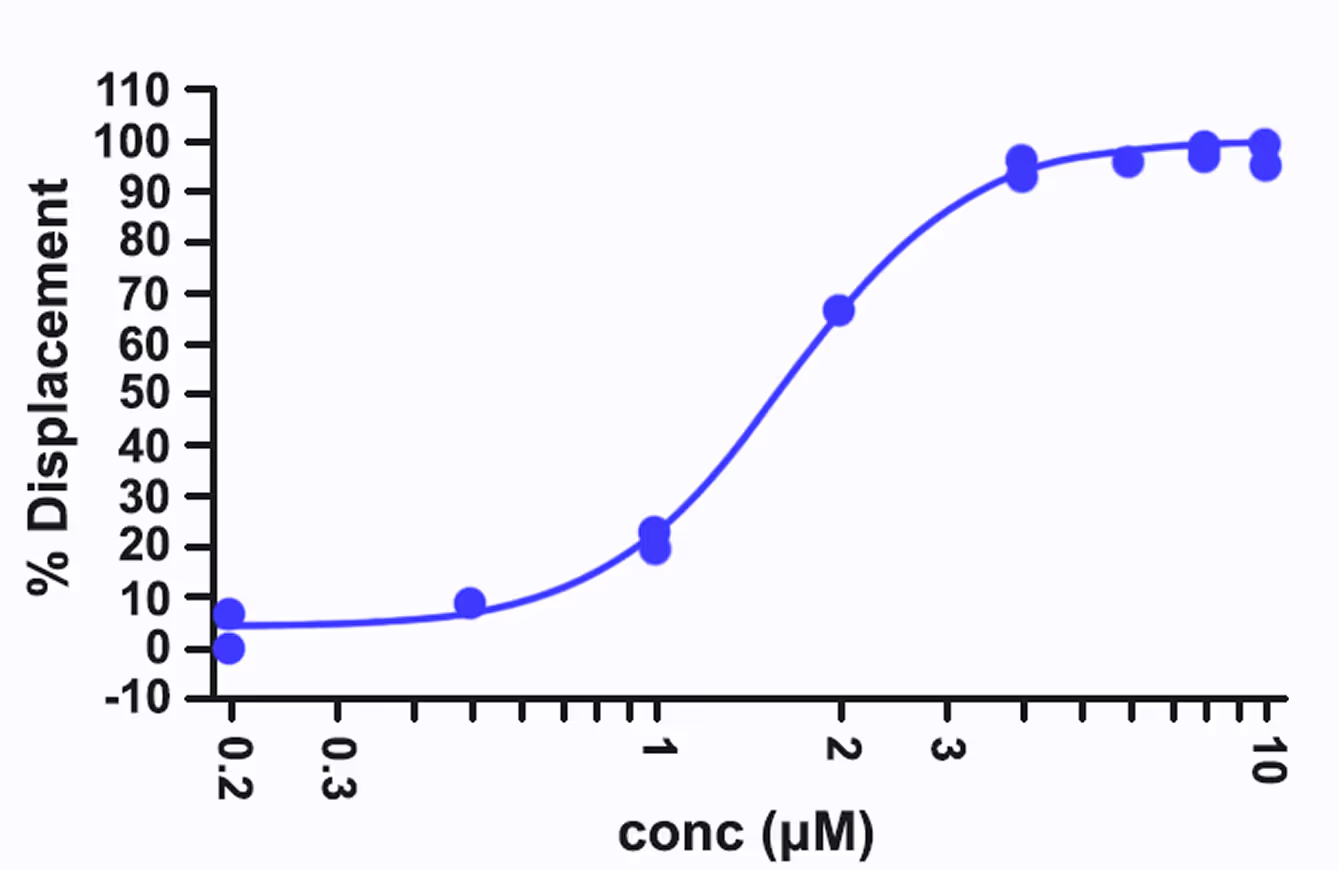
*Dose-response curve of the hit compound
with IC50 ~1.6 μM
with IC50 ~1.6 μM