~8M
compounds
screened
~12%
hit rate with 97 candidates
3
binding pockets
targeted
<10 μM
IC50 of 32 best hit compounds
0.35 μM
dissociation constant of the top compound
01/ Background
- Target is a heterodimeric receptor from the integrin family.
- It is a promising target in immunological disorders and cancers.
- There were no binding sites for small molecules known.
- Also, no potent small molecule binders were known.
- Target is often deemed undruggable by small molecules.
- The goal is to design a small molecule PPI disruptor.
02/ Methodology
- Conformational changes predicted with crystal structures using AI-driven MD simulations.
- Binding pockets identified within dynamic conformations by proprietary AI model.
- Virtual screening of 8M stock library.
- AI-docking with target-specific rescoring using ArtiDock.
- In vitro ligand displacement assay performed to validate hit candidates’ ability to compete with a known ligand for binding.
- SPR analysis performed to measure binding kinetics and affinity (dissociation constant — Kd).
03/ Results
- Identified 97 hits out of ~1,200 hit candidates, resulting in 12% hit rate.
- Identified 4 binding pockets, including one cryptic.
- Successfully targeted 3 pockets - Pocket I (41 hits), Pocket II (40 hits), and Pocket III (16 hits).
- 32 hit compounds with IC50 < 10 μM.
- The top hit shows the highest affinity (Kd = 0.35 μM).
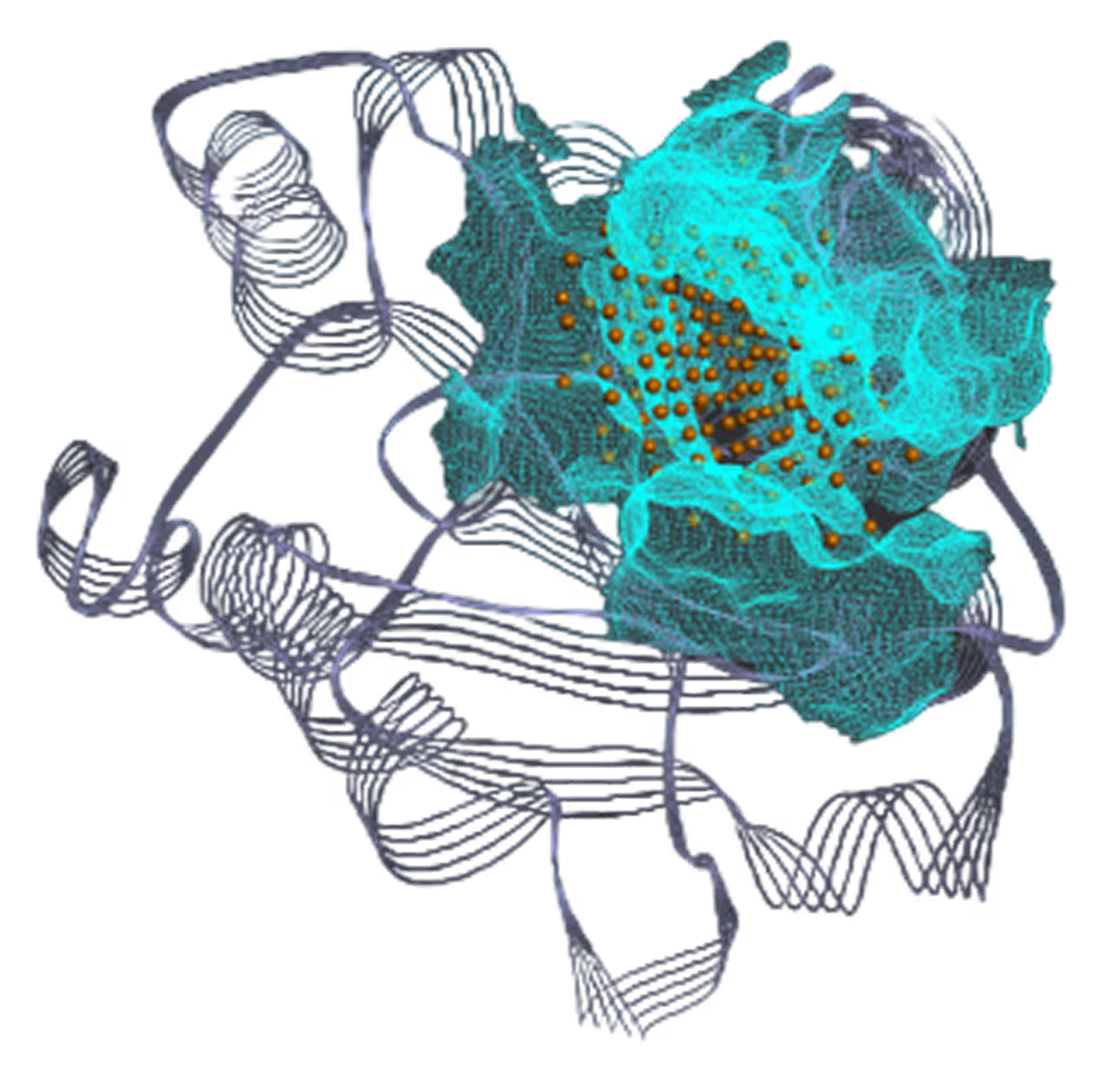
*Pocket I visualization
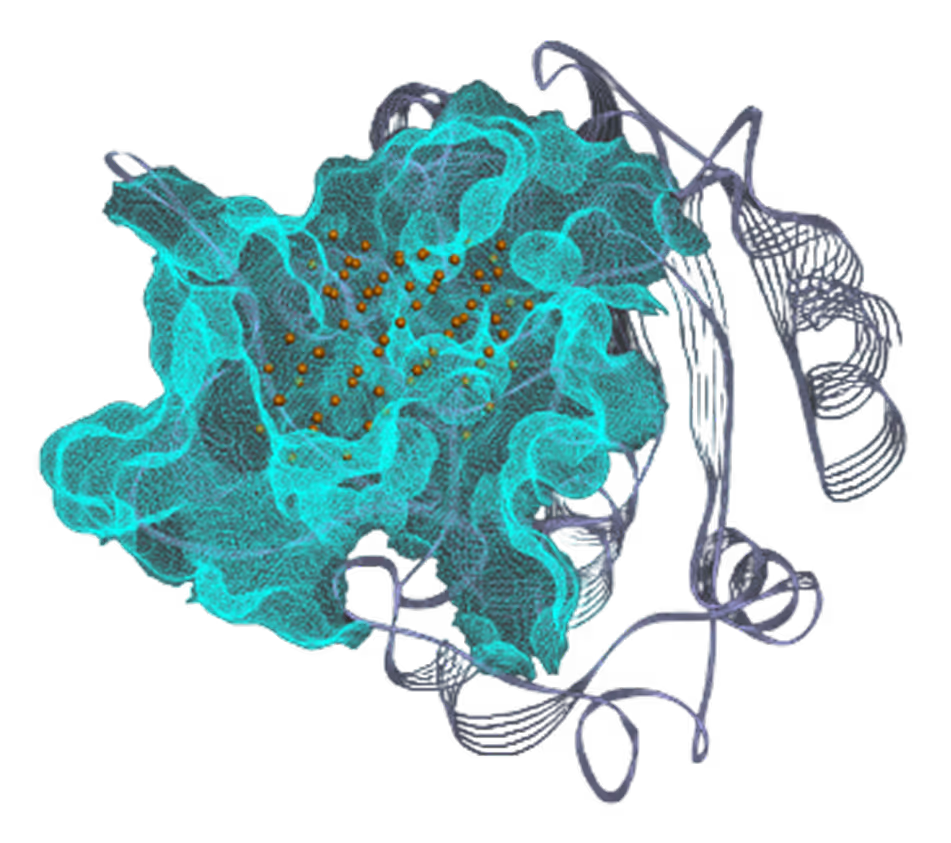
*Pocket II visualization
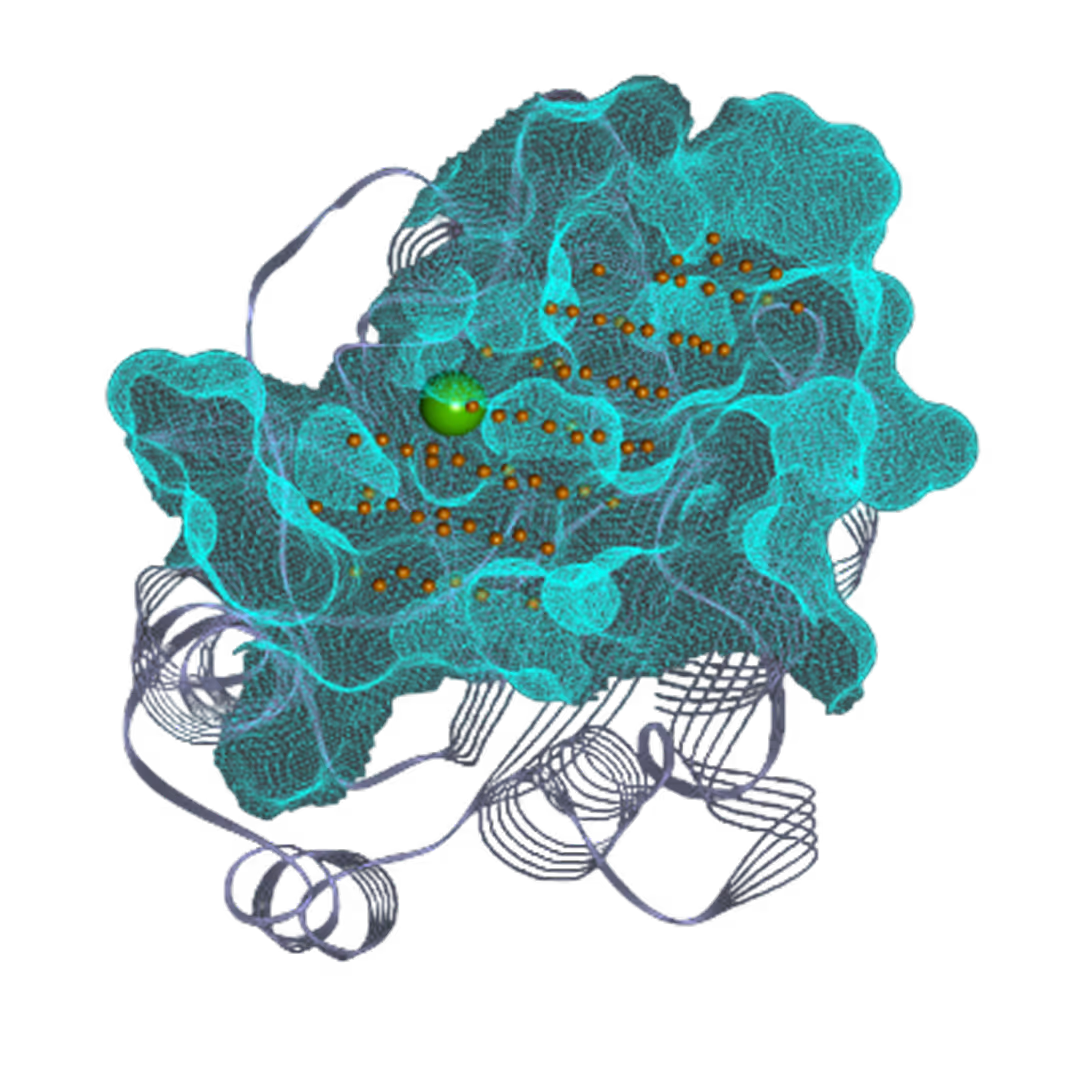
*Pocket III visualization

