AI vs Physics: Outperformance in Protein–Ligand Docking

AI vs Physics: Outperformance in Protein–Ligand Docking
PARTNERSHIP
Announcement



Summary
Full Text
We evaluate docking approaches on the PoseX benchmark and outline a hybrid workflow that combines AI-generated poses with brief physics-based minimization for optimal accuracy and throughput. Physics-based methods are robust but slow and often struggle with metal-coordinating sites or structured water. AI docking predicts plausible poses in seconds and captures interaction patterns that are hard to encode with hand-crafted scoring.
Overall Outperformance
Standardized benchmarks of experimentally solved complexes allow direct pose-accuracy comparisons. We used the PoseX dataset, reflecting real discovery challenges.

Pure AI engines such as ArtiDock and Uni-Mol yield a higher fraction of correct poses than classic force-field methods like AutoDock Vina or Glide. Accuracy improves further when ArtiDock’s pose is followed by a brief physics-based refinement (Vina or UFF), adding little compute. Co-folding models that dock while predicting the pocket are useful in some cases but currently trail both specialized AI docking and physics-based methods.
Performance in Difficult Areas
Sites with metal ions, cofactors, or structured water are the hardest; generic parameterizations can miss coordination geometry and directional hydrogen bonding, causing misplaced poses.
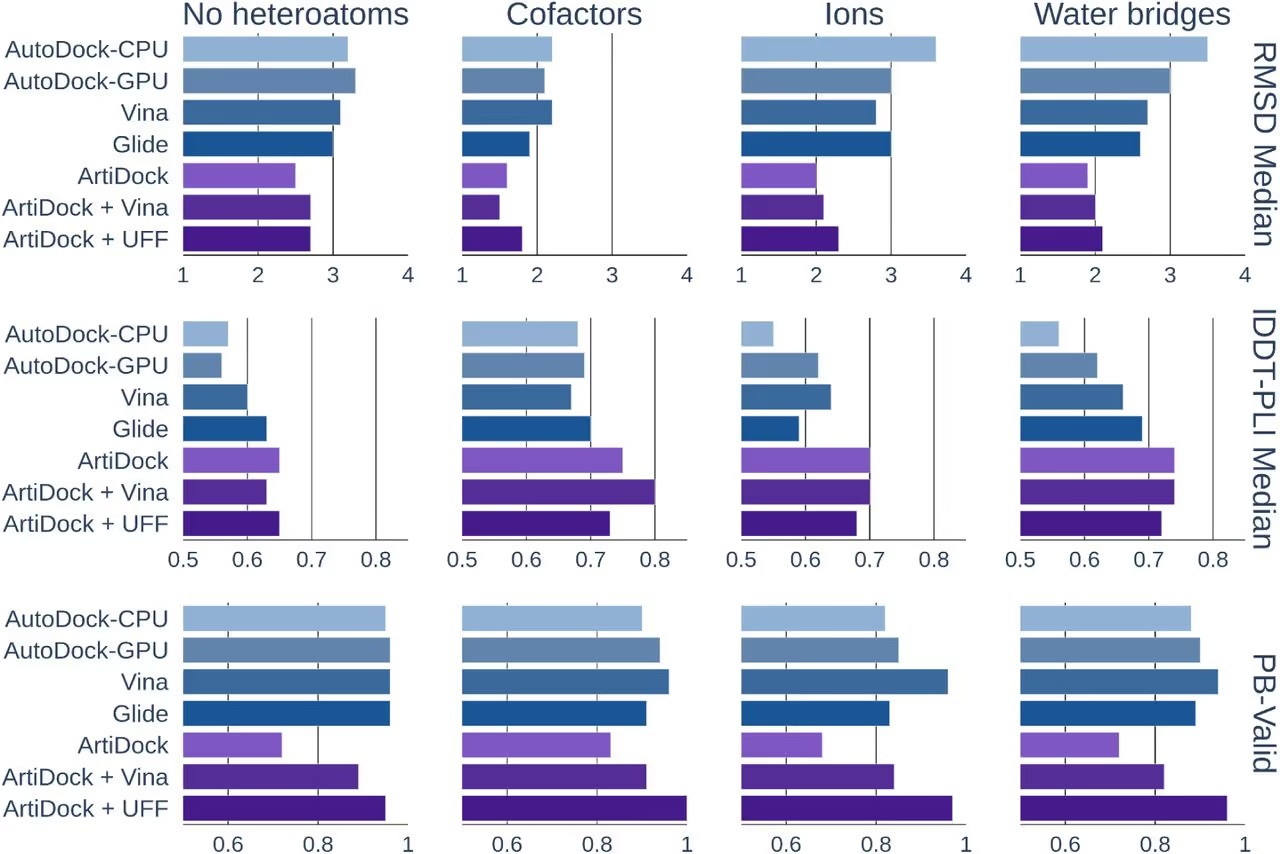
ArtiDock shows lower median RMSD and better recovery of true contacts across difficult-site categories. A short minimization may slightly increase RMSD due to subtle shifts but typically boosts chemical validity by restoring key interactions.
How It Works: ArtiDock Architecture

ArtiDock encodes the ligand and pocket as compact graphs with essential chemical and spatial features. A lightweight network learns an inter-graph distance matrix that describes how the ligand should fit. A fast alignment then converts the predicted distances into a 3D pose, bypassing exhaustive search.
Conclusion
A hybrid AI–physics workflow delivers precise poses in seconds, scales to large libraries, and performs especially well in metal/cofactor/water-rich pockets, accelerating decision-making in drug discovery.

