Identification of dual-targeting Mycobacterium tuberculosis aminoacyl-tRNA synthetase inhibitors using machine learning
Identification of dual-targeting Mycobacterium tuberculosis aminoacyl-tRNA synthetase inhibitors using machine learning
PARTNERSHIP
Announcement



Summary
Full Text
Mycobacterium tuberculosis, the bacterium responsible for tuberculosis (TB), is a major global health threat. According to WHO Global tuberculosis report, in 2021, tuberculosis was the second deadliest infectious killer after COVID-19. Each year, TB infects millions of people and causes over a million deaths worldwide. While TB is a treatable and curable disease, the emergence of multidrug-resistant TB (MDR-TB) has complicated efforts to control and eliminate the disease. MDR-TB is defined as TB that is resistant to the most effective first-line antibiotics. The emergence and spread of MDR-TB is a serious concern because it makes TB more difficult and expensive to treat, and it increases the risk of the disease becoming untreatable in the near future.
Recently the CTO and Head of Computational chemistry of Receptor.AI, Dr. Starosyla, co-authored the scientific paper devoted to the search of novel dual-targeting drugs against multidrug-resistant tuberculosis.One of the promising strategies for overcoming the resistance is designing drugs which target multiple bacterial proteins at once. This slows down the adaptation of pathogens to such drugs and gives them longer life in clinics until the resistance will eventually form.
The aminoacyl-tRNA synthetases are commonly used as antibiotic targets. These enzymes play a crucial role in the process of protein synthesis. They are responsible for attaching specific amino acids to their corresponding transfer RNA (tRNA) molecules, a process called aminoacylation. Bacterial aminoacyl-tRNA synthetases are significantly different from their human counterparts, which makes them promising selective drug targets.
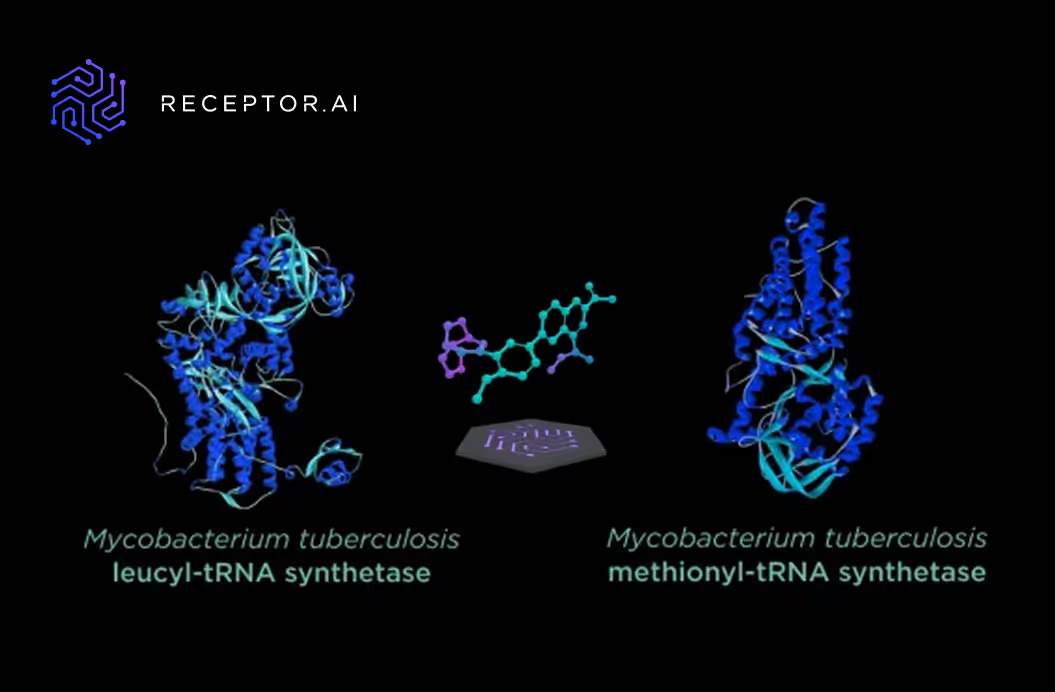
Two enzymes were chosen as the targets: leucyl-tRNA synthetase (LeuRS) and methionyl-tRNA synthetase (MetRS). The probability of simultaneous mutation in both enzymes, which leads to drug resistance, is negligible thus, the drugs which act on both of them should be successful in the long run.
The neural network models were built to search for prospective dual-targeting drugs. The training and test sets were generated based on the known bacterial LeuRS and MetRS inhibitors taken from the ChEMBL database. 92 LeuRS and 252 MetRS known inhibitors were processed and divided into training and testing datasets. Since the amount of data was severely limited, there was a risk of getting the “narrow-minded” models, which work poorly with unknown ligand structures. To avoid this, data augmentation was performed by calculating up to 100 conformers for each known compound and using their 3D fingerprints for model training.
The obtained artificial neural network models for LeuRS and MetRS were used for virtual screening of the OTAVA compound collection containing about 250,000 compounds in order to select the compounds for biochemical testing. According to the results of machine learning, we have picked 200 compounds for investigation of their inhibitory activity simultaneously toward M. tuberculosis MetRS and LeuRS in an aminoacylation assay. The compounds of interest were defined as those decreasing the activity of enzymes by more than 50% at the concentration of 100 μM in aminoacylation assay. Among the tested compounds, we identified 24 MetRS inhibitors, 10 LeuRS inhibitors and 6 prospective dual-targeted inhibitors.
The best compounds found by the AI model were subject to molecular docking to prove their binding to the active site of each of the aminoacyl-tRNA synthetases.
The best dual activity was observed for 2-(quinolin-2-ylsulfanyl)-acetamide. This compound was subject to further modification, and 14 derivatives were synthesised and tested in order to improve its activity. One of the derivatives, called 10a, has shown superior activity. It inhibits MetRS and LeuRS with IC50 values of 33 μM and 23.9 μM, respectively.
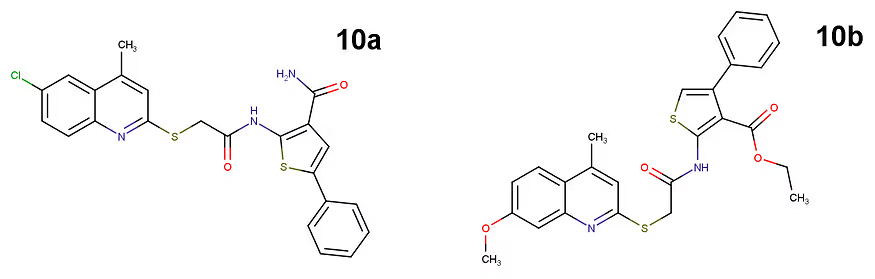
Molecular docking confirmed that 10a binds to the active sites of both enzymes and forms multiple favorable interactions with the crucial amino acids.
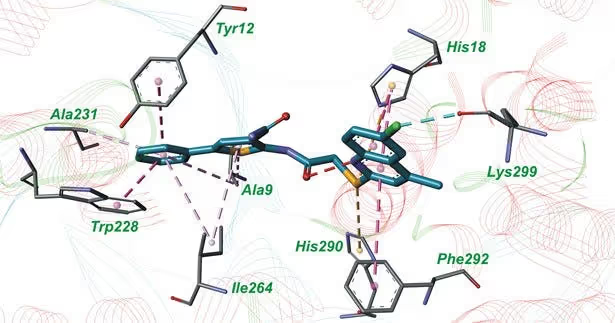
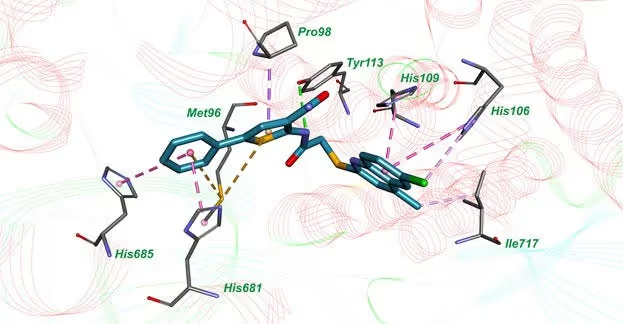
This study demonstrates that even a rather simple and straightforward machine learning approach could be useful in identifying dual-targeted inhibitors of M. tuberculosis MetRS and LeuRS. The most active compound is suitable for further chemical optimisation and biological research. It has the potential to be developed into a drug candidate against multi-drug-resistant tuberculosis.
This paper is academic research which was started before Receptor.AI launched its drug discovery platform. Now our capabilities leave the approaches used in this work many miles behind. We are excited about applying the full power of our current AI technologies to the challenging task of fighting antibiotic resistance, and the previous experience of our team gives us an advantage in such endeavours.

